Probably the scariest cancer to get is brain cancer because of its precarious location and often advanced grade of disease when diagnosed. However, earlier detection could be on the way after a research team from Hong Kong and mainland China showed that an affordable technology used in a few research labs is more sensitive and accurate in detecting the presence of the most common type of brain tumour from biopsies than the older technology currently adopted by hospitals. They hope their proof of concept will also lead towards the eventual development of a test using the less invasive method of a blood sample to detect brain tumours in people who currently show no or minimum symptoms.
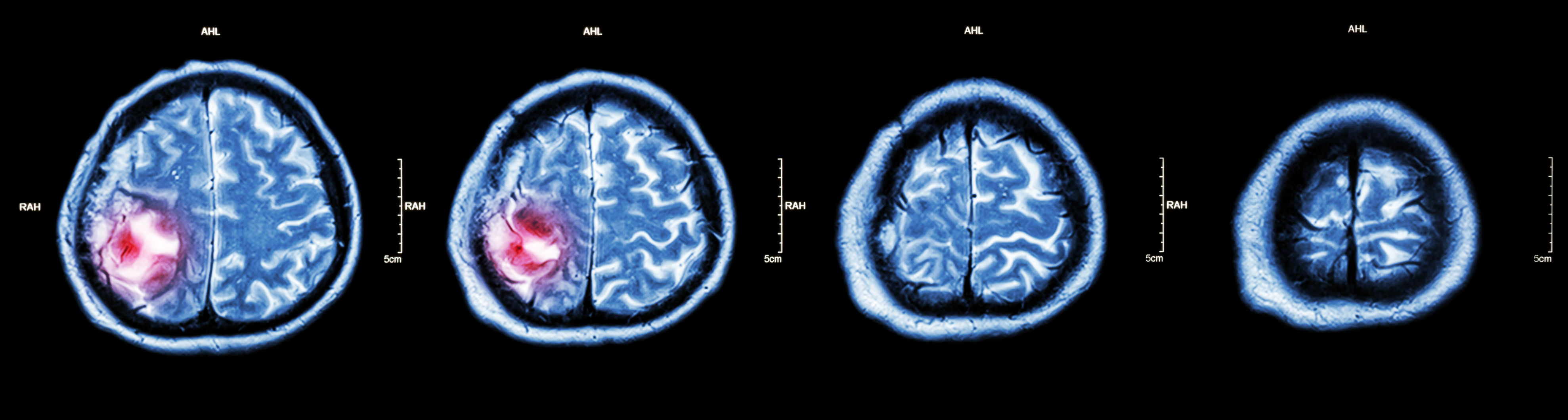
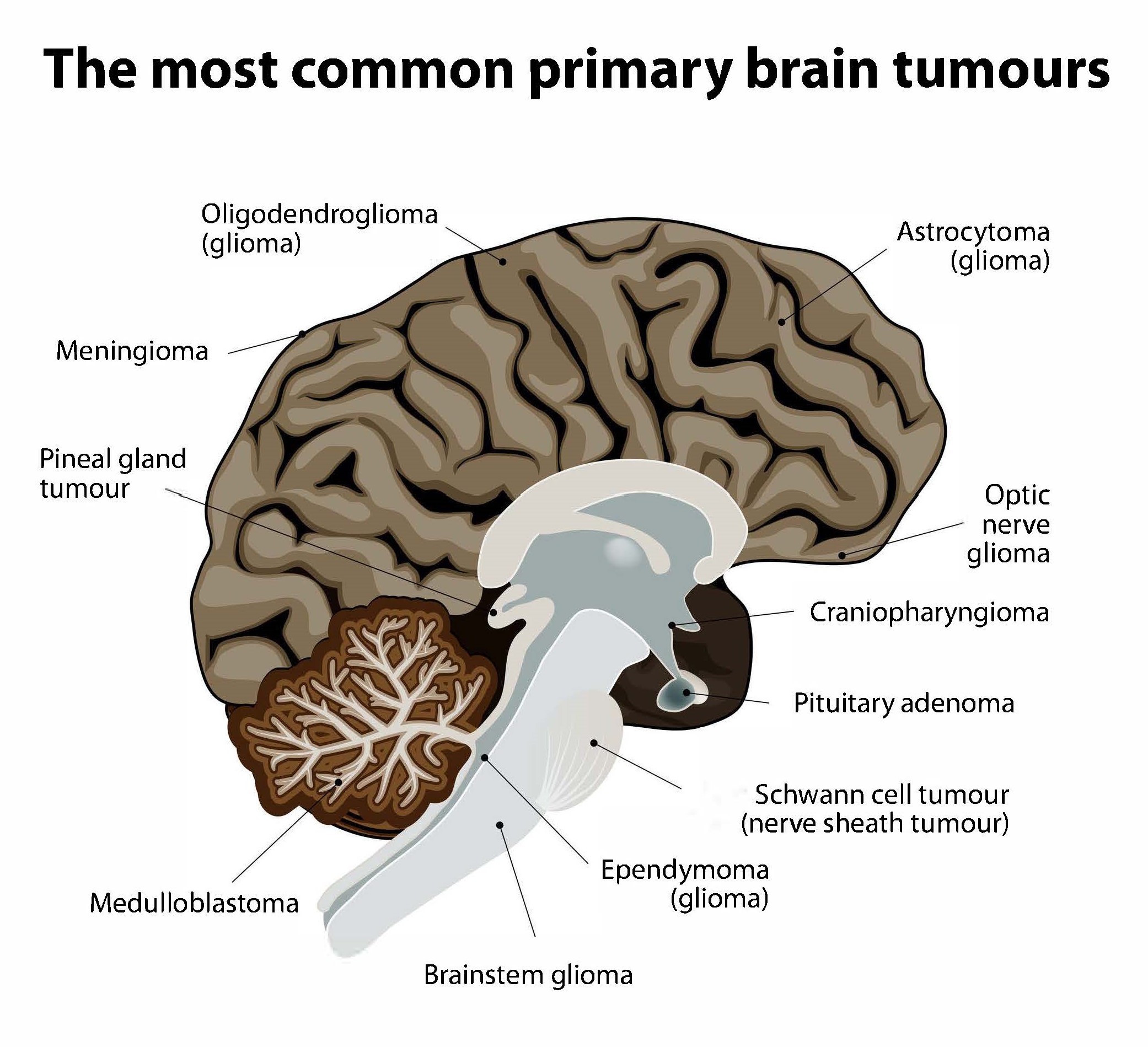
Non-cancerous (benign) and cancerous (malignant) primary brain tumours begin in the brain, while secondary brain tumours are malignant tumours that have spread to the brain from a cancerous part of the body. About 30%-40% of all brain tumours are gliomas, a type of primary brain and spinal cord tumour that starts out as benign. Some 80% of all cancerous primary brain tumours begin as gliomas, and between 15%-20% of all primary brain tumours are glioblastomas, which were low-grade gliomas that had worsened to grade IV, the most aggressive kind of cancerous brain tumour. Many glioblastoma patients survive only for a year or so after diagnosis. However, lower grade gliomas are also potentially life-threatening if they are not removed.
In recent years, doctors have been able to take advantage of the finding that mistakes or “mutations” in IDH1 and IDH2 genes have been detected in more than 70% of low-grade gliomas and some glioblastomas. In particular, the IDH1(R132H) mutation has been found quite frequently and early on in the development of gliomas, making it an effective diagnostic and predictive biomarker or ‘natural indicator’ of the presence of a glioma.
Many hospitals use quantitative real-time polymerase chain reaction technology (qPCR or qRT-PCR) to detect and extrapolate the number of IDH1 mutations in a biopsied sample of a brain tumour. Since biopsied samples may contain only a small number of copies of the target mutation, qPCR works by first generating multiple copies of the known sequence of genetic code containing the target mutation and using “probe” molecules tagged with fluorescence or another marker that allows copies to be detected. During this “amplification” in little “wells” in plastic “plates” or in small plastic assay tubes, the machine displays a mathematical curve in real time that shows a logarithmic relationship between the amount of fluorescence or other probe marker detected and the number of amplification cycles needed to produce those amounts. The curve is used to derive the number of copies of the mutation in the sample.
The research team, which included Associate Professor Dr Tony To Shing-shun of PolyU’s Department of Health Technology and Informatics, wanted to see if the newer droplet digital polymerase chain reaction technology (ddPCR) could be more sensitive in detecting and calculating the number of IDH1 mutations. The team noted previous studies had demonstrated that ddPCR could be successfully applied for different genetic and biomedical purposes, such as detecting a particular mutation in breast cancer biopsies which resulted in an ineffective type of treatment being abandoned and the adoption of other treatments that did not require a biopsy.
Similar to qPCR, ddPCR first generates multiple copies of the target mutation by polymerase chain reaction and using probes with fluorescence or another marker. However, ddPCR differs from qPCR in the way it quantifies. qPCR uses plastic wells or tubes, which, owing to the size of the containers, limit the number of possible “reaction chambers” to a few hundred that can potentially yield data. qPCR also requires users to repeat the experiment a few times using the sample diluted a known number of times to derive the mathematical curve since too much or too little of the target mutation or reagent chemicals and other substances can skew the results. In contrast, ddPCR generates tens of thousands of discrete droplets of an almost identical nanolitre size containing a mixture of the sample, reagents and stabilising chemicals. ddPCR then analyses the droplets to determine if at least 1 copy of the mutation is present or not in each droplet. Poisson statistics is then used to calculate the total number of copies of the mutation in the sample. Since each droplet is informative, the sensitivity and accuracy of ddPCR are much greater than that of qPCR.
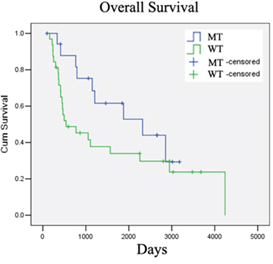
To study whether ddPCR could indeed be a more sensitive diagnostic and monitoring tool for the IDH1 mutation, Dr To and his collaborators obtained consent from 62 mainland Chinese patients with grades II to IV of glioma to test biopsied samples from their brain tumour specimens using ddPCR, qPCR, and Sanger direct sequencing as a benchmark. Although Sanger sequencing is usually not used in hospitals and cannot quantify, it is currently the gold standard for discovering the exact sequence of bases — and therefore the locations of mutations — in genetic material.

Sensitivity is usually defined as how likely a test will indicate all the actual instances where the target mutation is present (“true positives”), while specificity is often regarded as how likely a test will indicate all the actual instances where the target mutation is absent (“true negatives”). Using the biopsied samples, the research team found that ddPCR was a more sensitive test for detecting the IDH1 mutation, with 100% sensitivity, while it was just 90.48% for qPCR. However, the specificity of ddPCR was lower than that of qPCR, with ddPCR scoring only 95.56% while qPCR attained 100%.
However, any potential diagnostic or screening tool for health care usage must also be reliable and cause as few false alarms as possible while not missing any actual instances, so the prevalence of the particular mutation in the target population has to be considered too. Positive predictive value is how likely a person who has been found by a test to have the mutation actually has the mutation, while negative predictive value is how likely a person who has been found by a test as not having the mutation actually does not have the mutation. The higher the prevalence of the mutation, the higher the positive predictive value and the lower the negative predictive value. Meanwhile, the lower prevalence, the lower the positive predictive value and the higher the negative predictive value.
The team found that the positive predictive value and the negative predictive value for ddPCR were 90.48% and 100%, respectively, and 100% and 95.56% for qPCR, respectively. This together with the sensitivity and specificity results implied that ddPCR would be better at predicting negative results and for screening a large number of people, while qPCR would be better at predicting positive results and for confirming the detection of the mutation. In addition, the cost of ddPCR is currently only slightly higher than that of qPCR and should therefore be affordable to health institutions.